Frontiers in Medicine 01 frontiersin.org
Case report: Local bacteriophage
therapy for fracture-related
infection with polymicrobial
multi-resistant bacteria: hydrogel
application and postoperative
phage analysis through
metagenomic sequencing
VolkerAlt
1†
, AndréGessner
2†
, MayaMerabishvili
3
,
FlorianHitzenbichler
4
, GopalaKrishnaMannala
1
,
DavidPeterho
2
, NikeWalter
1
, Jean-PaulPirnay
3
,
AndreasHiergeist
2‡
and MarkusRupp
1
*
‡
1
Department of Trauma Surgery, University Hospital Regensburg, Regensburg, Germany,
2
Institute of
Clinical Microbiology and Hygiene, University Hospital Regensburg, Regensburg, Germany,
3
Laboratory for Molecular and Cellular Technology (LabMCT), Queen Astrid Military Hospital, Brussels,
Belgium,
4
Department of Infection Prevention and Infectious Diseases, University Hospital
Regensburg, Regensburg, Germany
Fracture-related infections can be challenging, particularly with concomitant
severe bone defects and multi-resistant microorganisms. We present a case
of a 42-year-old patient with a fracture-related infection following a war
injury from a gunshot, resulting in a 12-cm subtrochanteric segmental bone
defect and the detection of four dierent multi-resistant Gram-negative
bacteria. Due to antibiotic drug resistance, treatment with bacteriophages was
considered. Phage susceptibility testing revealed the activity of a commercially
available bacteriophage cocktail (Intesti bacteriophage, Eliava Institute, Tbilisi,
Georgia). This phage cocktail was included in a modified two-stage Masquelet
technique. During the first intervention, the bone was debrided and samples for
microbiological and phage testing were harvested. The indwelling intramedullary
rod was removed, and the bone defect was filled with a PMMA spacer loaded
with colistin and the bone stabilized with a plate. During the second procedure,
the PMMA spacer was removed and a silver-coated angular stable plate was
implanted. The bone defect was filled with a fibular autograft and allograft
cancellous bone chips. At the end of the procedure, the Intesti bacteriophage
cocktail was injected into a DAC hydrogel and this bacteriophage hydrogel
composite was then put onto the angular stable plate. Postoperatively the wound
fluid was collected over 72 h, and high-throughput metagenomic sequencing
was performed. This showed a time-dependent release of the bacteriophages
in the wound fluid, with a relatively high concentration after 12 h, decreasing to
DNA copies of 0 after 72 h. Furthermore, wehave assessed the release of phages
from DAC gel and the eect of DAC gel on the phages in vitro. The results
showed a stable and rapid release of phages from the DAC gel (~1×10
3
PFU/mL).
The clinical course of the patient showed no relapse of the infection with good
bone consolidation of the bone defect after 1 year without the need for any
surgical revision. To the best of our knowledge, this is the first case that shows
the detection of bacteriophage DNA copies by high-throughput metagenomics
OPEN ACCESS
EDITED BY
William Calero-Cáceres,
Technical University of Ambato, Ecuador
REVIEWED BY
Seth Commichaux,
UnitedStates Food and Drug Administration,
UnitedStates
Swapnil Ganesh Sanmukh,
Université Clermont Auvergne, France
*CORRESPONDENCE
Markus Rupp
markus.rupp@ukr.de
†
These authors share first authorship
‡
These authors share senior authorship
RECEIVED 06 May 2024
ACCEPTED 20 June 2024
PUBLISHED 12 July 2024
CITATION
Alt V, Gessner A, Merabishvili M,
Hitzenbichler F, Mannala GK, Peterho D,
Walter N, Pirnay J-P, Hiergeist A and
Rupp M (2024) Case report: Local
bacteriophage therapy for fracture-related
infection with polymicrobial multi-resistant
bacteria: hydrogel application and
postoperative phage analysis through
metagenomic sequencing.
Front. Med. 11:1428432.
doi: 10.3389/fmed.2024.1428432
COPYRIGHT
© 2024 Alt, Gessner, Merabishvili,
Hitzenbichler, Mannala, Peterho, Walter,
Pirnay, Hiergeist and Rupp. This is an
open-access article distributed under the
terms of the Creative Commons Attribution
License (CC BY). The use, distribution or
reproduction in other forums is permitted,
provided the original author(s) and the
copyright owner(s) are credited and that the
original publication in this journal is cited, in
accordance with accepted academic
practice. No use, distribution or reproduction
is permitted which does not comply with
these terms.
TYPE Case Report
PUBLISHED 12 July 2024
DOI 10.3389/fmed.2024.1428432
Alt et al. 10.3389/fmed.2024.1428432
Frontiers in Medicine 02 frontiersin.org
sequencing in a patient with a complex fracture-related infection. Successful
treatment of this case encourages further investigation of bacteriophage
therapy in patients with complex bone and joint infections.
KEYWORDS
bacteriophage, fracture-related infection, metagenomic, Masquelet, hydrogel
1 Introduction
e treatment of fracture-related infections (FRI) is challenging.
Particularly, the involvement of severe bone defects and multi-
resistant bacteria frequently leads to complex situations. e treatment
of FRIs mainly consists of two key aspects: First, control of the
infection and causative microorganisms must beachieved. Second,
bone reconstruction with subsequent successful bone healing needs
to beobtained (1).
Both local and systemic antibiotic therapy are hallmarks of
fracture-related infection treatment. However, there are oen only
limited antibiotic treatment options when multi-drug resistant
bacteria are involved. An alternative or complementary approach for
the treatment of complex infections with multi-drug resistant bacteria
is bacteriophages (short phages). e bactericidal eect of phages was
rst identied more than 100 years ago by Felix d’Herelle (2). Since
then, phages have been studied and used mainly in the former Soviet
Union during the second half of the 20th century. In recent years,
bacteriophage therapy has also gained interest in the Western world
due to the rise of multi- and pan-drug-resistant bacteria, including
dicult-to-treat bone and joint infections (3). However, there are only
a few published case series or case reports on the treatment of
musculoskeletal infections with bacteriophages (4). A recent review
identied 33 bone and joint infection cases treated with phages, out of
which notably 29 (87%) achieved either microbiological or clinical
success. Furthermore, 8 of the 33 cases (24%) experienced mild and
temporary adverse eects, but there were no reports of serious
complications (5). In addition, underlining the ecacy of phage
therapy in resolving dicult-to-treat infections, Pirnay etal. have just
recently published clinical outcomes of their rst 100 cases treated
with personalized phage therapy. Bone infections were the three most
common indications. Overall, in 77.2% of targeted infections, patients
experienced clinical improvement. In 61.3% of infections where
relevant bacteriological follow-up data were available, eradication of
the targeted bacteria was observed (6). Ongoing clinical trials aim to
rene phage application mode and advance our understanding of how
to eectively manage bone infections. Currently there are a few clinical
trials registered, which already entered the phase of recruiting patients
for phage treatment.
Hitherto, however, the optimal administration of bacteriophages
has not yet been elucidated. Phages can beadministered intravenously
or locally. Dierent approaches with varying quantity, frequency, and
type of application have been described to date (4, 7). Recently, Ferry
et al. published a case report with the use of hydrogel as carrier
material for the application of bacteriophages in a patient with a mega
prosthetic hip infection (8). is study showed a rapid release of the
phages from the DAC hydrogel in an in vitro setting. However, no in
vivo data for the release of the phages in the patient were presented.
erefore, in this study, wepresent a clinical case involving the
loading of bacteriophages into a hydrogel in an FRI. Additionally,
weassess the in vivo release kinetics of the phages from the hydrogel
in the patient’s wound using high-throughput metagenomic
sequencing-based characterization of the administered bacteriophage
cocktail, followed by real-time PCR quantication of selected
bacteriophages in the drainage uid.
2 Case description
A 42-year-old man suered a war injury during the Russian war in
Ukraine. He had a history of a gunshot and a blast injury to his
proximal le femur 8 months ago. Hefurther sustained an open head
injury and a fracture of the 3rd metacarpal of his le hand. e injury
of the proximal femur also included a lesion of the sciatic nerve, mainly
aecting the peroneal part. e patient has been treated in Ukraine
with several debridement and irrigation procedures of the le femur,
along with the application of a ring xator. Multiple revision surgeries
for debridement, and irrigation of local antibiotics resulted in a 12-cm-
wide segmental subtrochanteric bone defect. Aer the transferal to
Germany, the external xator was removed, and the implantation of an
antibiotic-coated intramedullary rod was performed. e
microbiological results of this revision surgery revealed several multi-
drug resistant pathogens such as multi-drug resistant (MDR)
Pseudomonas aeruginosa, MDR Proteus mirabilis, MDR Klebsiella
pneumoniae, MDR Escherichia coli, and vancomycin-resistant
Enterococcus faecium. A combination therapy with daptomycin,
cederocol, and fosfomycin was established. Aer presentation in our
department, a modied two-staged Masquelet procedure was
performed with the removal of the antibiotic-coated rod and
debridement of the subtrochanteric bone defect. Microbiological
analysis of the tissue samples harvested during the rst Masquelet
procedure surgery revealed Cutibacterium acnes, which was sensitive
to penicillin G, vancomycin, and clindamycin. In addition,
S. epidermidis susceptible to vancomycin, rifampin, and cotrimoxazole
was detected. e previously cultivated multi-resistant Gram-negative
bacteria could not bedetected. e 12-cm segmental bone defect was
managed with a daptomycin- and colistin-loaded polymethyl
methacrylate (PMMA) spacer (80 g Copal
®
(Heraeus Medical,
Germany) + 6 Mio. I.E colistin/CMS and 2 g daptomycin), and proximal
femur was stabilized with an angular stable plate (NCB, Zimmer
Biomet, USA). Systemic antibiotic therapy was initially performed with
fosfomycin 3x5g, colistin (CMS) 3x3MioIU, and daptomycin 1x1g and
administered until nal surgery. Cultures of the initially evidenced
MDR pathogens were sent to the Queen Astrid Military Hospital,
Brussels, Belgium, for phage susceptibility testing. Aer testing the
available phages, the best possible option for additional treatment
Alt et al. 10.3389/fmed.2024.1428432
Frontiers in Medicine 03 frontiersin.org
seemed Intesti bacteriophage cocktail (Eliava Institute, Tbilisi,
Georgia). e phage cocktail then was used with a hydrogel carrier to
improve the release kinetics of this composite biomaterial, as described
in a recently published case by Ferry etal. (8). Aer 92 days during
which phages were tested and wound healing was achieved, the nal
surgery for bone defect reconstruction was performed. e patient was
informed about the intended compassionate use of phages according
to §37 of the Helsinki Declaration, and written consent was obtained
(9). e indwelling PMMA spacer and the angular stable plate were
removed, and the bone ends were debrided again. Five tissue samples
were sent for microbiological analysis, which revealed no detectable
pathogen. Perioperative antibiotic therapy with daptomycin and
meropenem i.v. was supplemented with colistin i.v. for 14 weeks
postoperatively and nished aer negative long-term incubation.
ere was no sign of persistent infection, and the decision was taken
to perform a bone reconstruction with an ipsilateral autologous bula
gra and allogenous cancellous bone within a nicely formed Masquelet
membrane. e subtrochanteric bone region was stabilized by a
combination of a long proximal femur nail (Gamma nail, Stryker,
USA) together with a silver-coated (HyProtect™ coating, Bio-Gate,
Germany) angular stable plate (NCB Zimmer Biomet, USA). e
angular stable plate was slightly bent in order to allow a high volume
of bone graing of the defect with a subsequent spindle-shaped callus
formation. e Defensive Antibacterial Coating (DAC
®
, Novagenit,
Italy) gel loaded with bacteriophage cocktail was applied onto the plate
before wound closure (Figure1).
Wound drainage uid and serum samples taken aer 12, 24, 48,
and 72 h were evaluated regarding the release of bacteriophages from
the hydrogel into the wound.
e patient was then followed up for 12 months. e wound
healed uneventfully, and there was no sign of reinfection, nor was
there a need for further surgical revision. X-rays and computed
tomography demonstrated sucient bone healing of the graed area,
achieving bone consolidation aer 12 months.
3 Materials and methods
3.1 Testing activity of phages on the patient
strains
e activity of phages was tested by spot-test using 100-fold
dilutions of Intesti bacteriophage cocktail (e Eliava IBMV),
individual phages, phages produced as active pharmaceutical
ingredients (API) available from the collection of Queen Astrid
Military Hospital, Brussels, Belgium. Intesti bacteriophage is a mix
of sterile ltrate of phage lysates active against Shigella spp.
(Shigella exneri serotype 1,2, 3, 4 and Shigella sonnei) (titer no less
than 10
5
mL
−1
), Salmonella spp. (S. patarype A, S. paratype B,
S. typhimurium, S. enteritidis, S. cholera suis, S. oranienburg) (titer
no less than 10
5
mL
−1
), dierent types of E. coli (titer no less than
10
5
mL
−1
), Proteus mirabilis and Proteus vulgaris (titer no less than
10
5
mL-
1
), Staphylococcus aureus (titer no less than 10
5
mL
−1
),
Pseudomonas aeruginosa (titer no less than 10
5
mL
−1
), and
Enterococcus spp. (titer no less than 10
5
mL
−1
). Inactive ingredients
are bacterial growth media, standard sodium saline, and
Chinazolin as a preservative. Phage APIs have quality control
certicates from Sciensano (formerly known as the Belgian
Scientic Institute of Public Health) and are ready-to-use
pharmaceutical ingredients to be used in magistral phage
preparations (10). Shortly, each phage was diluted 100 times in
DPBS and 10 μL of each dilution was spotted on the lawn of
bacterial strains made with semi-solid 0.6% lysogenic broth (LB)
agar as a second layer on the surface of LB agar in square Petri
plates. e starting concentration of each phage except Intesti
bacteriophage was 10
9
plaque-forming units per milliliter (PFU/
mL). e results were recorded aer overnight incubation at
32°C. Individual plaques were counted where visible and eciency
of plating (EOP) was dened as a ratio of the titers of phage on the
test strain and the host strain.
3.2 High-throughput metagenomic
sequencing
In order to analyze the composition of the Intesti
bacteriophage cocktail, metagenomic DNA was extracted from
2 mL of the original suspension before administration. DNA was
extracted following the protocol of Shkoporov etal. (11). Briefly,
viral particles were precipitated by treatment with polyethylene
glycol followed by DNAse and RNAse digestion of free nucleic
acids. DNA was extracted from the precipitate containing
bacteriophages using phenol/chloroform extraction. Sequencing
was performed on the original bacteriophage suspension. For this
purpose, DNA was fragmented to ~150 bp using the M-220
focused ultrasonicator (Covaris, Woburn MA, USA) followed by
two-sided size selection with MagSi-NGSPREP-PLUS beads
(magtivio, Nuth, The Netherlands) using a DNA to bead ratio of
1:1.1 and 1:0.6. The Ion Plus Fragment Library Kit (Thermo
Fisher Scientific, Whatman, MA, USA) was used to prepare the
sequencing libraries. The final library was quantified using the Ion
Library TaqMan™ Quantitation Kit (Thermo Fisher Scientific,
Whatman, MA, USA) and diluted to a concentration of 70
pM. The sequencing library was amplified using the Ion 550™ Kit
on an Ion Chef instrument and loaded onto a 550 chip. High-
throughput sequencing was performed on an Ion GeneStudio™
S5 Plus instrument, resulting in a total of 3.9 million reads with
an average read length of 161 bp.
3.3 Analysis of viral metagenomic
sequencing data
Raw sequencing reads were obtained from the Torrent Suite
v5.18.1 instrument soware and then quality-ltered using
Trimmomatic v0.9 (12). Sequencing adapters were removed using
cutadapt v4.0 (13). Reads mapping to the human GRCh37 reference
genome were removed using bowtie2 v.2.5.0 and samtools v1.17 (14,
15). Filtered reads were assembled using megahit v1.2.9. e VIRify
pipeline was used to detect and classify viral contigs (16). Sourmash
v4.8.0 was used for viral taxonomic classication of viral contigs by
MinHash sketching and searching against the viral genbank 2022.03
database (17). Here, a k-mer length of 31 and a scaled value of 1,000
were used for calculating signatures from assembled viral contigs.
Default parameters were used for taxonomic classication using the
Sourmash tax algorithm.
Alt et al. 10.3389/fmed.2024.1428432
Frontiers in Medicine 04 frontiersin.org
3.4 Quantification of bacteriophage DNA in
drainage fluid
In order to quantify viral bacteriophage genomes in wound
drainage uid or blood, a total of 10 mL sample material was taken
at ve dierent time points aer surgery: immediately aer surgery,
12 h post-surgery, and on days 1, 2, and 3 aer surgery. DNA
extraction was executed as described above. e two largest contigs,
which matched to Proteus and Pseudomonas bacteriophage
genomes, were selected to quantify bacteriophage DNA copies from
the original bacteriophage suspension and drainage uids collected
at dierent times: First 12 h, 12 h, and 1, 2, and 3 days post-
operatively. Primers S12_k141_521_f (5’-TATCTATCCCTCTCC
CGCCG-3′) and S12_k141_521_r (5’-GTTGAAGATAACGCCG
ACGC-3′) were used to detect the viral contig VC_521
(Pseudomonas phage KPP10). Primers S12_k141_755_f (5’-AGTGT
GTACAGAGCCAGTGC-3′) and S12_k141_755_r (5’-GCGGTAT
CACCAGCTAGCAT-5′) were designed to target a fragment of viral
contig VC_755 (Proteus phage vB_PmiP_RS1pmA). Fragments
cloned into the pJET 1.2 cloning vector (ermo Fisher Scientic,
Whatman, MA, USA) were used to quantify bacteriophage DNA
from nucleic acid extracts on a LightCycler 480 II using the
LightCycler
®
480 SYBR Green IMaster (Roche Molecular Sciences,
Rotkreuz, CH).
3.5 The release of pages and DAC gel eect
on the phages in vitro
To analyze the release of the phages from DAC gel and the eect
of the DAC gel on the phages, wehave incubated the DAC gel loaded
with Intesti bacteriophage in PBS at 37°C. e activity of the phages
was tested against the E. coli at dierent time points (0 h, 0.5 h, 1 h, 2 h,
4 h, and 6 h) using double-layer agar method.
4 Results
All ve strains of dierent species were tested against the phage
collection of Queen Astrid Military Hospital, Brussels, Belgium, and
the Intesti bacteriophage cocktail produced by the Eliava IBMV. In
particular, P. aeruginosa strain was tested against 6 phage APIs and 18
individual phages. Only four individual phages proved to beactive
with EOP in the range of 1.0–0.01; three active phages were
representatives of the Pakpunavirus species. e nomenclature
aliation of the fourth phage was not known.
E. faecium strain was tested against 1 API and 4 individual
phages. K. pneumoniae strain was tested against 27 individual
phages. E. coli tested was tested against 5 individual phages. None
of the APIs or individual phages showed any activity on the above
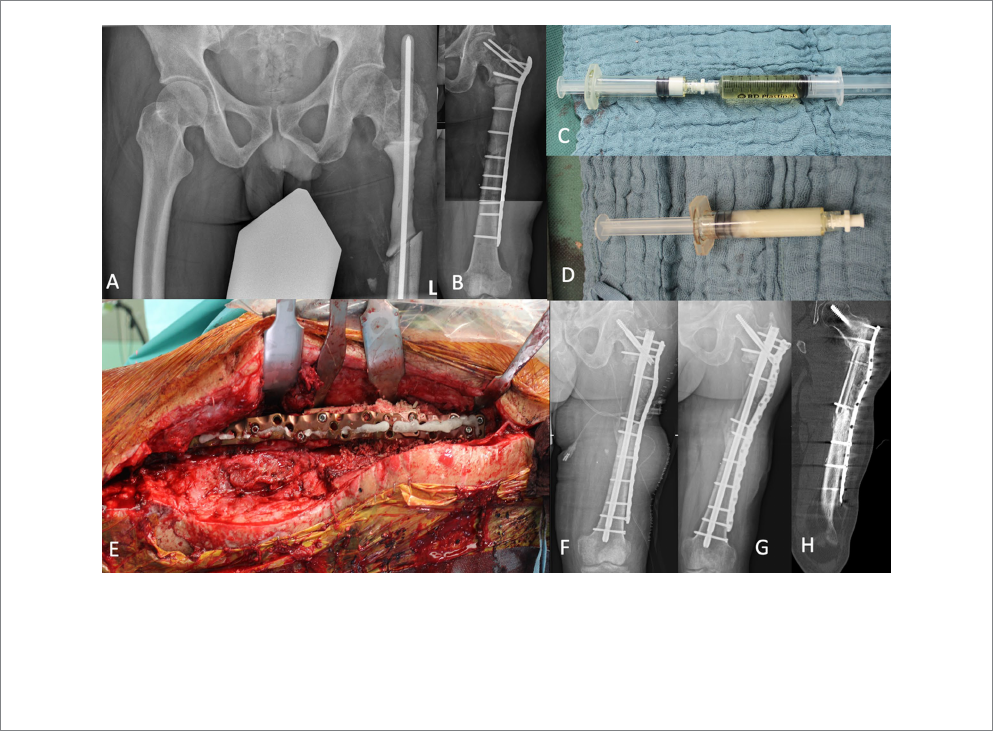
FIGURE1
Subtrochanteric segmental bone defect of the left femur was temporarily stabilized with an intramedullary custom-made PMMA bone cement-coated
rod (A). After removal of the rod, a PMMA spacer with colistin was added for dead space management and the bone stabilized with an angular stable
plate (B). The second stage of the Masquelet procedure was performed with an intramedullary nailing and augmentation plate. During bone defect
reconstruction phages were applied to the DAC® gel (C,D). The DAC gel loaded with Intesti bacteriophage was finally placed on the silver-coated
plate (E). Postoperative X-ray demonstrates the bone defect filled with autograft fibula graft and allogenic cancellous bone (F). After 12 months,
sucient bone healing was confirmed by performing X-ray (G) and computed tomography (H).

Alt et al. 10.3389/fmed.2024.1428432
Frontiers in Medicine 05 frontiersin.org
three strains. P. mirabilis was tested only with an Intesti
bacteriophage cocktail. Intesti bacteriophage showed activity
only against the E. coli strain. e titer of the phage(s) in the
cocktail active against the E. coli strain of the patient was dened as
2 ×10
3
PFU/mL.
As no active phage API was available against the bacterial strains and
the patient needed to betreated urgently, it was decided to start treatment
with Intesti bacteriophage, as the only available option at that time.
4.1 In vivo assessment of bacteriophage
release from hydrogel
In order to characterize the composition of bacteriophage genomes
within the Intesti bacteriophage suspension, nucleic acids were
extracted directly from the administered bacteriophage suspension
using whole shotgun metagenomic sequencing. A total of 511,078 (72,7
Mbp) raw sequencing reads were assembled to 149 contigs
(N50 = 9,914 bp). Of these, 77 could beidentied as viral contigs using
the VIRify pipeline. e taxonomic assignment resulted in 10 dierent
bacteriophage genera, which comprised Seunavirus, Saphexavirus,
Nankokuvirus, Kayvirus, Tunavirus, Hanrivervirus, Chivirus,
Bruynoghevirus, Teseptimavirus, and unclassied Casjensviridae.
Further k-mer-based searching against the GenBank nt database-
matched bacteriophages that target bacterial hosts Staphylococcus,
Enterococcus, Proteus, Shigella, Salmonella, Escherichia coli, and
Pseudomonas (Figure2). is was fully in line with the list of bacterial
hosts in the Intesti bacteriophage package insert.
To measure the release of bacteriophages from hydrogel in
drainage samples, specic real-time quantitative PCR protocols were
developed. ese protocols were based on two selected metagenomic
viral contigs, identied through k-mer-based searching against the
GenBank nt database, which presumably encodes bacteriophages
targeting bacterial species of the genus Proteus and Pseudomonas.
Genomic copies of bacteriophage from the Intesti bacteriophage
cocktail prior to administration and from drainage samples taken at 5
dierent time points aer surgery were quantied by qPCR:
immediately aer surgery, 12 h post-surgery, and on days 1, 2, and 3
aer surgery. e analyses revealed that bacteriophages were released
from the hydrogel within the rst 3 days following surgery (Figure3).
No DNA of these two bacteriophages was detected by qPCR in the
blood samples taken at the same time points.
4.2 In vitro assessment of bacteriophage
release from DAC hydrogel
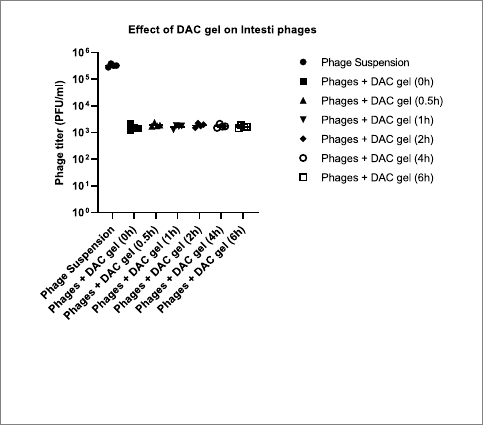
e release kinetics of the DAC hydrogel injected with Intesti
bacteriophages showed the rapid release of the phages from hydrogel
aer incubation in PBS. ere were no dierences in the release of
bacteriophages with the time progress (0 h-6 h) at 37°C incubation.
e released phage titer was between 1.6 × 10
3
and 1.9 × 10
3
PFU/mL,
which indicates a fairly consistent release rate of the phages from the
DAC gel (Figure4).
5 Discussion
In the present case of a Ukrainian soldier wounded in the Russian-
Ukrainian war, a combination of local bacteriophage therapy with
established therapeutic procedures achieved infection eradication and
bone reconstruction of the injured leg despite chronic osteomyelitis
caused by multiple MDR bacteria. In vitro analysis evidenced a
consistent release of phage from the DAC hydrogel. Furthermore,
phage analysis conducted in the postoperative course demonstrated
the activity of phages in the wound area during the rst postoperative
days, with no systemic spread of phages through the blood detectable
FIGURE2
Composition of the Intesti bacteriophage cocktail as revealed by
whole shotgun metagenomic sequencing. Viral contigs were
analyzed by k-mer-based searching against the GenBank nt
database.
FIGURE3
Quantification of bacteriophage release by real-time PCR targeting
viral contigs encoding a Proteus and Pseudomonas targeting
bacteriophage in postoperative drainage fluid as well as in Intesti
bacteriophage suspension (viral copies per mL). Samples were
collected at 5 dierent time points: immediately after surgery, 12 h
post-surgery, and on days 1, 2, and 3. The time point of surgery is
marked with a dashed line.
Alt et al. 10.3389/fmed.2024.1428432
Frontiers in Medicine 06 frontiersin.org
within the initial 3 postoperative days. No side eects of phage therapy
were observed.
In orthopedic trauma surgical treatment, the local application of
phages appears promising. However, the optimal form of application,
required quantities, and the frequency of phage application remain
unclear (4). While some authors advise the use of a draining device
for the administration of phages into the wound cavities, the potential
of the hydrogel as a phage delivery system has been highlighted (6,
18). In vitro and in vivo results of the present case indicate that
application via a hydrogel may bea possible and practical solution.
ese ndings are in line with a previously published case reporting a
salvage therapy in a patient with an infected knee megaprosthesis. e
in vitro testing of the impact of the hydrogel on phage activity showed
a rapid release and also stable titers for at least 6 h indicating
compatibility (8). e present case shows that this innovative approach
is also feasible for the treatment of bone infection and that phages
delivered via hydrogel may oer a promising strategy by targeting
specic bacterial strains. Based on these results, localized and
sustained delivery of phages to the infection site could
bedemonstrated. Furthermore, it can beassumed that the ecacy of
phage therapy may beenhanced by maximizing contact time between
phages and bacteria compared to rinsing the wound cavities with
phages. Another potential implication involves the versatility of
hydrogel delivery systems, which enable the incorporation of other
therapeutic agents, such as antibiotics for a synergistic approach.
Metagenomic sequencing is a powerful tool for identifying and
characterizing bacteriophages. However, its analytical sensitivity poses
challenges in detecting low-abundance bacteriophages directly from
the original clinical sample material (19). Diluting eects are
particularly important for high-volume sample material such as
wound drainage uid or blood. Additionally, the complexity of these
samples, combined with the presence of host DNA, can mask
bacteriophages. erefore, weused specic real-time quantitative PCR
(qPCR) protocols to conrm the presence of phages, ensuring
sensitive detection of bacteriophages. Reduction in bacterial counts
and the stability of phage titers as well as qPCR protocols are
commonly employed to conrm the presence and activity of phages
post-release and from hydrogels (20). ese ndings are in line with
other studies that detected the release of bacteriophages from
hydrogels for several days (21). Nevertheless, detection will depend on
the sensitivity of the detection method and the sample material.
Furthermore, the development and standardization of phage cocktails
pose several challenges. is study highlights the need for rigorous
characterization to ensure the safety and ecacy of these cocktails.
e case presentation also highlights practical weaknesses in the
current therapeutic approach and reveals challenges to beaddressed. (1)
In three performed revision surgeries, dierent communities of
antibiotic-resistant microbes were observed in routine diagnostics
conducted through culture in each operation. is can oen beobserved
in bone and joint infections (22, 23). However, the therapeutic
consequences for antibiotic and alternative therapies, such as phage
application, still need to bedetermined. Very high local antibiotic doses
for MDR bacteria may lead to the initial disappearance of MDR bacteria
and eventual infection eradication (24). (2) Bacteriophage testing on the
identied MDR bacteria did not yield the initially desired results, except
for the Georgian phage cocktail, which demonstrated ecacy against
MDR E. coli. (3) e time elapsed until obtaining results was substantial.
While a 6-week interval between operations is typically required for a
two-stage Masquelet procedure, in this case, the therapeutic interval of
92 days is more than twice as long. erefore, the testing and production
of suitable phages must besignicantly optimized.
6 Conclusion
In conclusion, this case demonstrates the potential of localized
phage therapy delivered via hydrogel in managing complex
fracture-related infections with multi-drug resistant bacteria and
severe bone defects. e integration of phage therapy with
established surgical procedures resulted in successful infection
eradication and bone reconstruction. High-throughput
metagenomic sequencing provided insights into bacteriophage
release dynamics in the wound environment, supporting the
feasibility of sustained delivery for several days aer surgery in a
clinical context. e case study highlights the fact that applying
phages had no negative side eects. It remains uncertain in the
present case whether bacteriophage application has eectively
contributed to preventing reinfection. Despite practical challenges,
including microbial variability and timely production processes,
phage therapy holds promise as a personalized approach for patients
with challenging bone infections.
Data availability statement
e raw data supporting the conclusions of this article will
bemade available by the authors, without undue reservation.
Ethics statement
Ethical approval was not required for the studies involving
humans because this case falls under the category of intended
compassionate use of phages according to the §37 of the Helsinki
Declaration. e studies were conducted in accordance with the local
legislation and institutional requirements. e participants provided
their written informed consent to participate in this study. Written
informed consent was obtained from the participant/patient(s) for the
publication of this case report.
FIGURE4
The release of bacteriophages from hydrogel and the impact of DAC
gel on phages. The scatter dot plot graph was created using
GraphPad Prism 9.5.
Alt et al. 10.3389/fmed.2024.1428432
Frontiers in Medicine 07 frontiersin.org
Author contributions
VA: Conceptualization, Data curation, Formal analysis, Investigation,
Methodology, Validation, Writing – original dra. AG:
Conceptualization, Data curation, Formal analysis, Investigation,
Methodology, Project administration, Validation, Writing – review &
editing. MM: Formal analysis, Investigation, Methodology, Validation,
Writing – review & editing. FH: Investigation, Methodology, Validation,
Writing – review & editing. GM: Data curation, Formal analysis,
Investigation, Methodology, Validation, Writing – review & editing. DP:
Formal analysis, Investigation, Methodology, Validation, Writing –
review & editing. NW: Conceptualization, Investigation, Validation,
Writing – review & editing. J-PP: Investigation, Methodology, Validation,
Writing – review & editing. AH: Formal analysis, Investigation,
Methodology, Validation, Writing – review & editing. MR:
Conceptualization, Data curation, Investigation, Methodology, Project
administration, Validation, Writing – original dra.
Funding
e author(s) declare that no nancial support was received for
the research, authorship, and/or publication of this article.
Acknowledgments
The authors thank Tristan Ferry, Hospices Civils de Lyon,
Lyon, France, for sharing his experiences and providing the
DAC gel.
Conflict of interest
e authors declare that the research was conducted in the
absence of any commercial or nancial relationships that could
beconstrued as a potential conict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors
and do not necessarily represent those of their aliated
organizations, or those of the publisher, the editors and the
reviewers. Any product that may beevaluated in this article, or
claim that may bemade by its manufacturer, is not guaranteed or
endorsed by the publisher.
References
1. Rupp M, Walter N, Bärtl S, Heyd R, Hitzenbichler F, Alt V. Fracture-related
infection—epidemiology, etiology, diagnosis, prevention, and treatment. Dtsch Arztebl
Int. (2023) 121:17–24. doi: 10.3238/arztebl.m2023.0233
2. viaLibri. (1917). BACTERIOPHAGE: Sur un microbe invisible antagoniste des
bacilles dysentériques (Comptes rendus hebdomadaires des séances de l’Académie des
Sciences. Available at: https://www.vialibri.net/years/books/8122883/1917-dherelles-
felix-bacteriophage-sur-un-microbe-invisible-antagoniste (Accessed February 5, 2024)
3. Wellcomecollection. (2014). Antimicrobial resistance: Tackling a crisis for the
health and wealth of nations/the review on antimicrobial resistance chaired by Jim
O’Neill. Available at: https://wellcomecollection.org/works/rdpck35v (Accessed
February 5, 2024)
4. Ferry T. A review of phage therapy for bone and joint infections. Methods Mol Biol.
(2024) 2734:207–35. doi: 10.1007/978-1-0716-3523-0_14
5. Suh GA, Ferry T, Abdel MP. Phage therapy as a novel therapeutic for the treatment
of bone and joint infections. Clin Infect Dis. (2023) 77:S407–15. doi: 10.1093/cid/ciad533
6. Pirnay J-P, Djebara S, Steurs G, Griselain J, Cochez C, de Soir S, et al. Personalized
bacteriophage therapy outcomes for 100 consecutive cases: a multicentre, multinational,
retrospective observational study. Nat Microbiol. (2024) 9:1434–53. doi: 10.1038/
s41564-024-01705-x
7. Briot T, Kolenda C, Ferry T, Medina M, Laurent F, Leboucher G, et al. Paving the
way for phage therapy using novel drug delivery approaches. J Control Release. (2022)
347:414–24. doi: 10.1016/j.jconrel.2022.05.021
8. Ferry T, Batailler C, Petitjean C, Chateau J, Fevre C, Forestier E, et al. e potential
innovative use of bacteriophages within the DAC
®
hydrogel to treat patients with knee
Megaprosthesis infection requiring debridement antibiotics and implant retention and
so tissue coverage as salvage therapy. Front Med (Lausanne). (2020) 7:342. doi: 10.3389/
fmed.2020.00342
9. e World Medical Association. WMA declaration of Helsinki – ethical principles
for medical research involving human subjects. Ferney-Voltaire: World Medical
Association (2022).
10. Pirnay J-P, Verbeken G, Ceyssens P-J, Huys I, de Vos D, Ameloot C, et al. e
Magistral phage. Viruses. (2018) 10:64. doi: 10.3390/v10020064
11. Shkoporov AN, Ryan FJ, Draper LA, Forde A, Stockdale SR, Daly KM, et al.
Reproducible protocols for metagenomic analysis of human faecal phageomes.
Microbiome. (2018) 6:68. doi: 10.1186/s40168-018-0446-z
12. Bolger AM, Lohse M, Usadel B. Trimmomatic: a exible trimmer for Illumina
sequence data. Bioinformatics. (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
13. Kechin A, Boyarskikh U, Kel A, Filipenko M. cutPrimers: a new tool for accurate
cutting of primers from reads of targeted next generation sequencing. J Comput Biol.
(2017) 24:1138–43. doi: 10.1089/cmb.2017.0096
14. Danecek P, Boneld JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve
years of SAMtools and BCFtools. Gigascience. (2021) 10:giab008. doi: 10.1093/
gigascience/giab008
15. Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods.
(2012) 9:357–9. doi: 10.1038/nmeth.1923
16. Rangel-Pineros G, Almeida A, Beracochea M, Sakharova E, Marz M, Reyes Muñoz
A, et al. VIRify: an integrated detection, annotation and taxonomic classication
pipeline using virus-specic protein prole hidden Markov models. PLoS Comput Biol.
(2023) 19:e1011422. doi: 10.1371/journal.pcbi.1011422
17. Pierce NT, Irber L, Reiter T, Brooks P, Brown CT. Large-scale sequence comparisons
with sourmash. F1000Res. (2019) 8:1006. doi: 10.12688/f1000research.19675.1
18. Kim HY, Chang RYK, Morales S, Chan H-K. Bacteriophage-delivering hydrogels:
current Progress in combating antibiotic resistant bacterial infection. Antibiotics. (2021)
10:130. doi: 10.3390/antibiotics10020130
19. Lewandowska DW, Zagordi O, Geissberger F-D, Kufner V, Schmutz S, Böni J, et al.
Optimization and validation of sample preparation for metagenomic sequencing of
viruses in clinical samples. Microbiome. (2017) 5:94. doi: 10.1186/s40168-017-0317-z
20. Li Y, Li X, Duan H, Yang KD, Ye JF. Advances and optimization strategies in
bacteriophage therapy for treating inammatory bowel disease. Front Immunol. (2024)
15:1398652. doi: 10.3389/mmu.2024.1398652
21. Kopač T, Lisac A, Mravljak R, Ručigaj A, Krajnc M, Podgornik A. Bacteriophage
delivery systems based on composite PolyHIPE/Nanocellulose hydrogel particles.
Polymers. (2021) 13:2648. doi: 10.3390/polym13162648
22. Rupp M, Kern S, Weber T, Menges TD, Schnettler R, Heiß C, et al. Polymicrobial
infections and microbial patterns in infected nonunions – a descriptive analysis of 42
cases. BMC Infect Dis. (2020) 20:667. doi: 10.1186/s12879-020-05386-9
23. Walker LC, Clement ND, Yapp LZ, Deehan DJ. Change in organism between rst-
and second-stage revision for periprosthetic joint infection of knee arthroplasty
independently associated with increased risk of failure. Bone Jt Open. (2023) 4:720–7.
doi: 10.1302/2633-1462.49.BJO-2023-0067.R1
24. Krajewski J, Bode-Böger SM, Tröger U, Martens-Lobenhoer J, Mulrooney T,
Mittelstädt H, et al. Successful treatment of extensively drug-resistant Pseudomonas
aeruginosa osteomyelitis using a colistin- and tobramycin-impregnated PMMA spacer.
Int J Antimicrob Agents. (2014) 44:363–6. doi: 10.1016/j.ijantimicag.2014.05.023
